New Update: This project is now accompanied by a published paper on bioRxiv, which benchmarks molecular dynamics simulations across various cloud platforms, with a special focus on optimizing Google Colab for scientific computing: https://www.biorxiv.org/content/10.1101/2024.11.14.623563v1
Molecular dynamics (MD) simulations are a powerful computational tool used in various fields of science, including chemistry, biology, and materials science. GROMACS is a widely used open-source molecular dynamics simulation software designed for biochemical molecules.
Molecular Dynamics simulations traditionally relied on costly on-premises infrastructure like supercomputers or clusters for High-Performance Computing (HPC). However, in recent years, there's been a significant shift towards utilizing computing resources provided by commercial cloud providers. This shift is driven by the flexibility and accessibility these platforms offer, irrespective of an organization's financial capacity.
Popular platforms such as Google Compute Engine and AWS now provide scalable computing infrastructure. Researchers can harness these cloud resources to conduct Molecular Dynamics simulations efficiently, without the constraints of traditional on-premises setups. This transition empowers researchers with greater flexibility and cost-effectiveness in their computational work. Monitoring and managing usage is crucial for optimizing costs, and cloud providers often offer tools and calculators to help users estimate expenses based on their specific requirements.This repository demonstrates how to use compute engine resources like AWS cloud computing and Google Compute Engine to accelerate molecular dynamics simulations, enabling researchers to tackle larger and more complex systems while optimizing costs and scalability.
For Google Compute Engine (Google Cloud): Instructions for Google Compute Engine
For AWS: Instructions for AWS
For Google Colab: Instructions for Colab
GitHub repository: Link

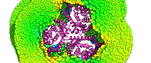
Molecular dynamics simulation on lysozyme: GIF animation without water & PNG with water
Image by Taner Karagöl, Programs: GROMACS & PyMOL
References/Further readings
Amazon Web Services. "Amazon EC2" Accessed February 22, 2024. https://aws.amazon.com/ec2/.
Kutzner, C., Kniep, C., Cherian, A., Nordstrom, L., Grubmüller, H., de Groot, B. L., & Gapsys, V. (2022). GROMACS in the Cloud: A Global Supercomputer to Speed Up Alchemical Drug Design. Journal of chemical information and modeling, 62(7), 1691–1711. https://doi.org/10.1021/acs.jcim.2c00044
For Benchmarks: Dept. of Theoretical and Computational Biophysics, Max Planck Institute for Multidisciplinary Sciences, Göttingen, https://www.mpinat.mpg.de/grubmueller/bench
Google. "Google Colaboratory" Accessed February 22, 2024. https://colab.research.google.com/.

Comments